What is the MIST Consortium?
The Mucosal Immunology Study Team (MIST) Consortium is an interdisciplinary group of scientists focused on immune defense mechanisms and immune regulation at mucosal surfaces. The over arching goal of the MIST program is to discover and define novel basic immune mechanisms, cells, mediators, and pathways that underly mucosal immune defense.
The GutPath Atlas helps to advance this mission by providing researchers with convenient access to a massive collection of single cell data from the small intestine across a wide range of well-controlled infection expeirments.
Who created the GutPath atlas?
The GutPath project is led by Daniel Beiting, a scientist who participates in the MIST Consortium and is faculty member at the University of Pennsylvania. All computational work to produce the atlas was carried out by Andrew Hart, PhD., with support from Rui Xiao, PhD. All infections and mouse work was carried out at the University of Pennsylvania with support from MIST Consortium faculty and their lab members.
How do I cite GutPath?
Please cite our preprint on BioRxiv
How is this work funded?
GutPath is funded through a grant to the MIST Consortium by the National Institutes of Allergy and Infectious Diseases (RFA-AI-15-023).
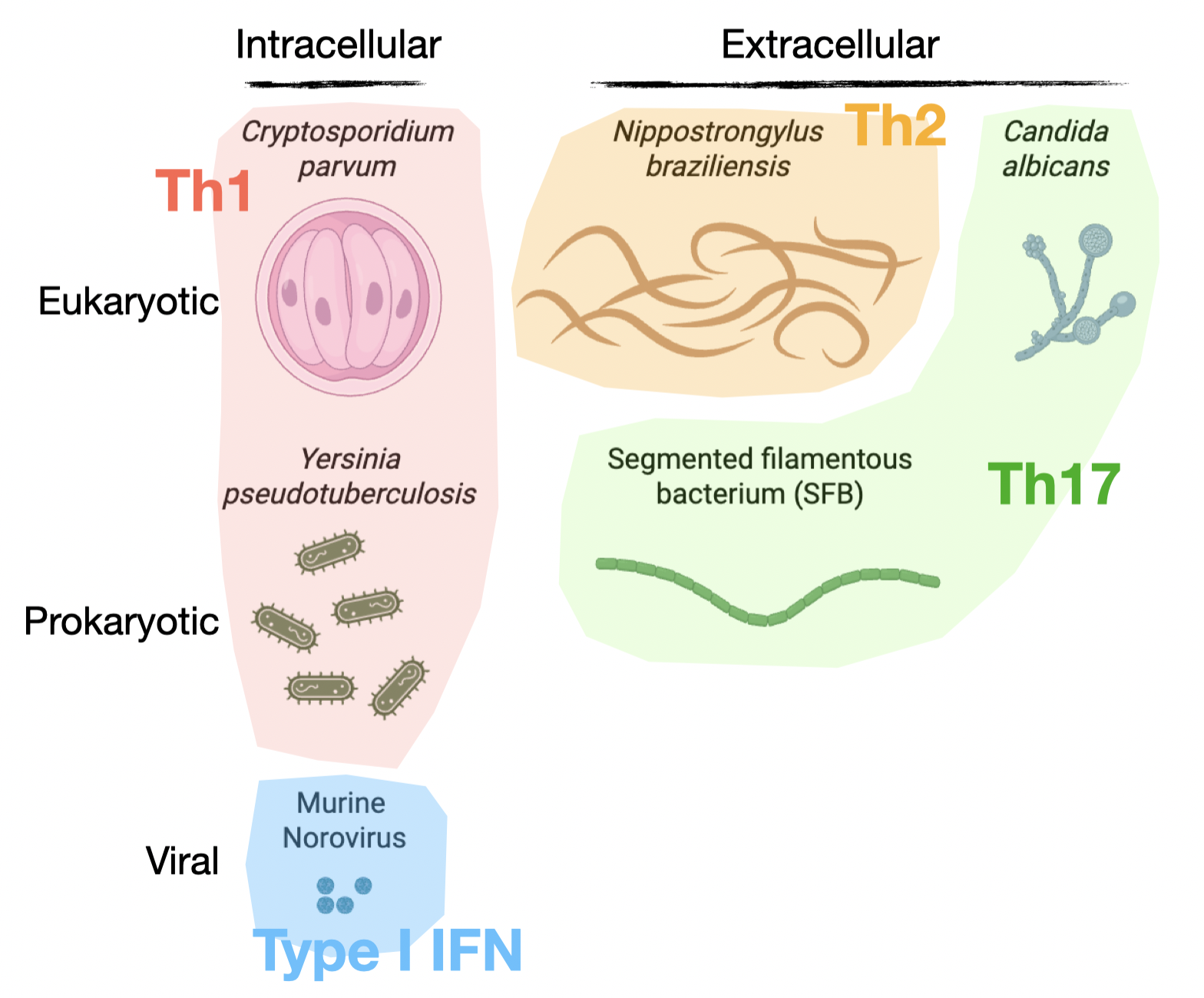
What pathogens were used in these studies?
Six pathogens were selected to represent a phylogenetically diverse group of microbes that infect the small intestine, including:
- the helmith parasite, Nippostrongylus brasiliensis
- the protozoan parasite, Cryptosporidium parvum, a leading cause of serious diarrheal disease worldwide
- the gram-negative bacterium, Yersinia pseudotuberculosis
- the gram-positive commensal, Segmented Filamentous Bacterium (SFB)
- the commensal fungus, Candida albicans
- Murine Norovirus
- Naive (uninfected) mice were used as controls
Notably, these pathogens elicit distinct mucosal immune responses, which means that our atlas includes an incredibly diverse array of cell types and states.
What technologies and reagents were used to generate GutPath?
All single cell transcriptomic data was produced using the 10X Genomics platform (Chromium X device) and the 3’ v4 GEM-X kit with feature barcoding, together with the BioLegend TotalSeq-B Universal Cocktail for mouse (~100 antibodies to surface epitopes; see full list here)
What mice were used for the infections?
GutPath was created using carefully controlled mouse studies in which all experiments were carried out in 6-10 week-old female mice sourced from The Jackson Laboratories. All mice were obtained from room EM03, which generates animals free of Segmented Filamentous Bacteria (SFB). Mice were maintained in the same room at the Veterinary School at the University of Pennsylvania. Infections were all carried out by oral gavage, except for Nippostrongylus brasiliensis, which was introduced subcutaenously.
How do I access the data?
You came to the right place! Just navigate back to the homepage by clicking on ‘GUTPATH’ in the upperleft of your browser. Each dataset is represented by a single card. Clicking on any card brings up a page that has a brief description and large “click here…” button. When you click the button you will launch a data exploration tool in a separate tab of your browser. This tool contains a single dataset loaded into the popular CELLXGENE software, which lets you carry out fairly detailed exploration and analysis.
In addition to our CELLXGENE implementation, on each dataset page you will also find a link to download a Seurat object containing the full annotated dataset that can be direclty loaded into your R environment. This allows you to use our cell type labels to annotate your own single cell or spatial transcriptomic data.
What about raw data and code?
Yep, we’ve got those too! You can find the raw data on the SRA, and all code is available in this Github repository.